Michael Deagen (Olsen Lab, MIT)
- BigSMILES: A Brief Background
- Drawing a Simple Polymer in Ketcher
- Defining Copolymer Architectures
- Further Specification of Copolymer Attributes
- Summary
- References
BigSMILES: A Brief Background
Informatics systems need a way to represent objects in a format amenable to storage and processing. In cheminformatics, molecules are represented through string-based line notations such as SMILES.1 While this notation works well for small molecules, polymers do not follow a well-defined structure. As a result, Lin et al. developed BigSMILES as a way to capture the stochastic (random or statistically determined) nature of polymers in a compact form familiar to practitioners in cheminformatics.2 The additional syntax and semantics within BigSMILES includes the notation of stochastic objects ({ }) whose connectivity is set via bonding descriptors ([<], [>], [$]). Otherwise, the definition of atoms and branching rules follow the SMILES syntax.
BigSMILES captures the chemistry of a polymer’s repeating units as well as the topology allowed based on the connectivity constraints of the bonding descriptors. The result is a compact representation of an ensemble that would be referred to as a given polymer, without over-specifying a single macromolecule within that ensemble. This post will cover how to use the chemical structure editor to draw polymers, as well as how to augment BigSMILES notation using the CRIPT data model.
Drawing a Simple Polymer in Ketcher
The CRIPT web application uses the Ketcher chemical structure editor3 for drawing 2D depictions of molecules or polymers, which are then converted into SMILES or BigSMILES line notation using custom code combined with the Javascript distribution of RDKit.4 To define a structural repeating unit (SRU), all atoms within the unit must be selected, as shown in the simple example of polyethylene (Fig. 1). When one or more SRUs are detected, the output line notation goes from SMILES to BigSMILES.

Defining Copolymer Architectures
Fig. 1 showed how to define a simple homopolymer in the Ketcher drawing tool to generate a BigSMILES identifier. BigSMILES also allows for the specification of copolymers, and the following sections will show how to draw these more advanced polymers so that all SRUs are converted into stochastic objects appropriately.
Block Copolymers
A diblock copolymer can be divided into two segments, where each segment contains a chain of repeating units of the same type. In this case, the BigSMILES line notation should follow the basic pattern { a }c{ b }, where a and b represent the respective structural repeating units defining each stochastic object and c is an optional segment bridging the blocks. To draw a block copolymer inside the Ketcher tool, each SRU must be defined separately (Fig. 2). The same process applies for other n-block copolymer topologies.

Statistical (or Alternating) Copolymers
A statistical (sometimes called “random”) copolymer contains a single stochastic object with a set of SRUs allowed within it, where bonding descriptors determine the connectivity constraints. In this case, the BigSMILES line notation should follow the pattern { a , b }, where a and b each represent a structural repeating unit with their own bonding descriptors.
To specify a statistical copolymer in the Ketcher drawing tool, the individual SRUs must be defined in addition to the SRU grouping all of them together (Fig. 3). The BigSMILES generator assumes head-to-tail configuration, but one can specify statistical coupling by replacing conjugate bonding descriptors [<] and [>] with the [$] bonding descriptor. To specify an alternating topology (ABABAB), each SRU should be surrounded by conjugate bonding descriptors of the same type (e.g., { [<]A[<] , [>]B[>] }).

Graft and Segmented Copolymers
Graft and segmented copolymers have a stochastic object within a stochastic object, either along a side chain (graft) or along the backbone (segmented). Here, the BigSMILES line notation contains a nested structure { a { b } }, where a represents the backbone and b represents the stochastic branch.
To specify a grafted polymer, the SRU of the backbone is defined independently of the SRU of the stochastic branch, and the hierarchical relationship between these SRUs is captured by the drawing tool (Fig. 4). To ensure that the branch SRU is fully contained, the branch atoms must all be highlighted along with the backbone SRU.

The order in which the backbone and nested SRUs are defined does not matter, as shown below in an example of segmented polyurethane (Fig. 5).

Star Polymers
A star polymer contains stochastic object “arms” extending from a central branching point, or seed. The BigSMILES notation in this case should follow a pattern such as { a } ... ({ b }) ... ({ c }), where the stochastic objects are contained within the SMILES branch notation ( ) and bonding descriptors capture the connectivity constraints. In the following simplified example, a star polymer is defined through SRUs extending from the seed molecule (Fig. 6).

These examples cover a large portion of polymer architectures supported by BigSMILES. Other polymer architectures (e.g., ladder polymers, hyperbranched networks) are also supported by BigSMILES notation, but certain limitations within Ketcher (specifically, the Molfile V2000 format used for parsing the molecule) prevent their specification through the current version of the chemical structure editor. For these more complex architectures, please refer to the BigSMILES documentation.5
Further Specification of Copolymer Attributes
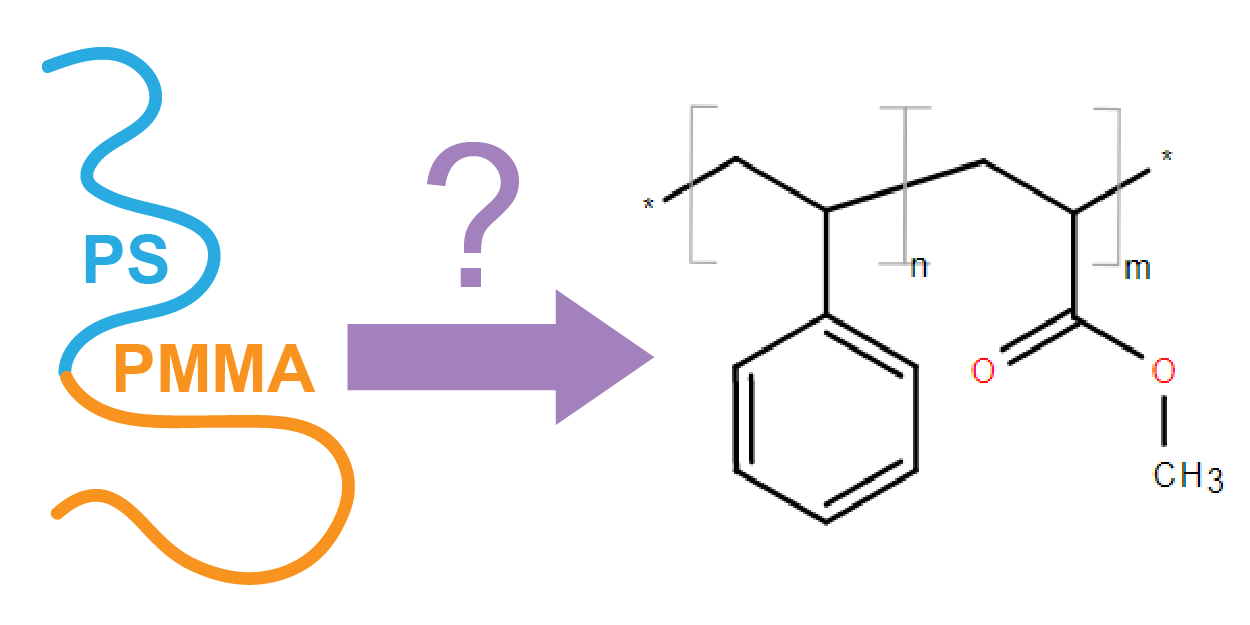
By design, BigSMILES notation does not capture instance-specific attributes of a polymer such as composition, polydispersity index, or molecular mass distribution. Instead, these additional data are captured within the CRIPT data model, which also enables the linking to a polymer’s detailed processing history as well as its characterization data. For example, the following JSON structure demonstrates a hypothetical specification of a diblock PS-b-PMMA polymer, where the composition of each substructure of the BigSMILES is specified with composition_fraction:
"identifiers": [
{
"key": "bigsmiles",
"value": "*{[<]COC(=O)C([>])C[<][>]}{[<][<]CC([>])c1ccccc1[>]}*"
}
],
"properties": [
{
"key": "composition_fraction",
"value": 0.6,
"structure": "{[<][<]CC([>])c1ccccc1[>]}"
},
{
"key": "composition_fraction",
"value": 0.4,
"structure": "{[<]COC(=O)C([>])C[<][>]}"
},
{
"key": "mw_n",
"value": 50,
"unit": "kg/mol"
}
]
Additional identifiers beyond SMILES and BigSMILES are offered through a controlled vocabulary (e.g. InChI, CAS, preferred name, lot number), which can be found in the CRIPT documentation.6 The controlled vocabulary of material properties can also be found in the documentation.7
Summary
This tutorial provided a brief introduction into how various polymers can be specified as BigSMILES line notation using the Ketcher chemical structure editor, as well as linking to instance-specific properties through the CRIPT data model. For information on how to programmatically access the CRIPT REST API using Python, please refer to the documentation for the Python SDK.8
References
- Weininger, David. “SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules.” Journal of chemical information and computer sciences 28.1 (1988): 31-36.
- Lin, Tzyy-Shyang, et al. “BigSMILES: a structurally-based line notation for describing macromolecules.” ACS central science 5.9 (2019): 1523-1531. DOI: 10.1021/acscentsci.9b00476.
- https://lifescience.opensource.epam.com/ketcher/
- https://www.rdkitjs.com/
- https://olsenlabmit.github.io/BigSMILES/docs/line_notation.html
- https://mycriptapp.org/vocab/material_identifier_key/
- https://mycriptapp.org/vocab/material_property_key/
- https://c-accel-cript.github.io/cript/